Research
nanomaterials -> neural interfaces -> biophysical theory
Current Interests
I study the statistical physics of biological ensembles and their interactions, focusing on RNA as a model system. I primarily work with Ofer Kimchi, Asst. Prof. of Mathematics at NYU's Courant Institute of Mathematical Sciences.
Biomolecular information theory
RNA both encodes information and interacts with other biomolecules following chemical rules. How does the statistical physics of RNA and its interactions impact the information it can contain and transfer, and vice versa? For example: given an ensemble of RNA molecules, each with a desired mate, on- vs off-target binding can be viewed as information propagation over a noisy channel. The dynamics of this process can be calculated using chemical kinetics. How does the efficiency of this information flow depend on the statistical physics of the interactions? Can we write down precise global metrics of information transfer (e.g. specificity, channel capacity, etc.) that capture important dynamics of a system?
Predicting RNA hybridization
Nucleic acids hybridize (e.g. stick together according to Watson-Crick base pairing rules, A-T/U, C-G) widely in biology— to replicate DNA/transcribe RNA, regulate gene expression, and in RNA therapies like the COVID vaccine—yet we lack a comprehensive theory that can predict the intensity and speed of hybridization between two arbitrary sequences, let alone ensembles of sequences.
I use tools from nonequilibrium statistical mechanics and machine learning to understand the principles of nucleic acid interactions. Working closely with experiment, we're able to infer hybridization between RNA and DNA strands from sequence and structure, potentially enabling precision RNA therapeutic design. We're also tackling the inverse problem—inferring RNA structure from binding data—which could dramatically simplify RNA folding prediction.
Past Themes
Neural interfaces (2016-2020)
In the Cui lab at Stanford Chemistry, I grew large magnetic nanorods inside human cells, with the goal of being able to open ion channels mechanically by applying a magnetic field that would torque the nanorods. This work, published in Nano Letters, is a step towards "magnetogenetics": remotely triggering neurons with magnetic fields.
I then spent a summer in college working at Paradromics, a leading brain machine interface company, where I developed a method to sharpen electrodes on the nanoscale to minimize tissue damage upon insertion into brain tissue. They still use this protocol, to my knowledge.
After college, I enrolled in the Harvard Applied Physics PhD (jointly with MIT) in 2020 to continue my work in neural interfaces. Not wanting to start grad school during the pandemic and excited by the idea that became Sora, I dropped out without starting, following in the footsteps of hip hop legend Lil Pump.
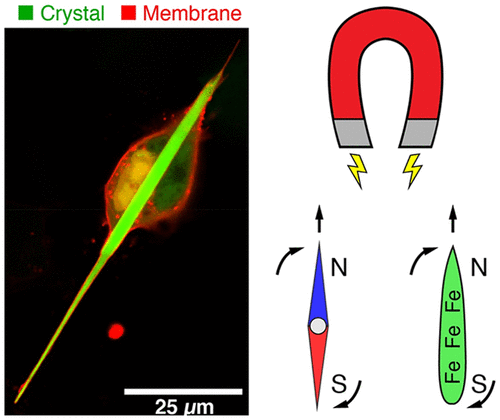
huge iron rods we grew inside human cells
Nanomaterials (2012-2016)
Working in the Tracy, Li, and Melechko labs at NC State University in high school, I created optically active nanomaterials for bioimaging applications, catalysts for biofuel generation, and scalable production methods for vertically-aligned carbon nanofibers used in drug delivery, respectively. I coauthored my first research paper, on the latter work, at age 14. I also spent the summer before college at RTI International developing stain-resistant textile coatings for a corporate client, using harmless silica nanoparticles instead of the industry standard "forever chemicals".
For my research work in high school, I was named a Semifinalist in the Intel Science Talent Search and a Regional Finalist in the now-defunct Siemens Competition.
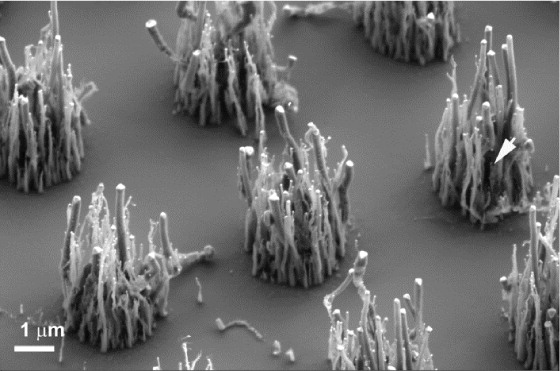
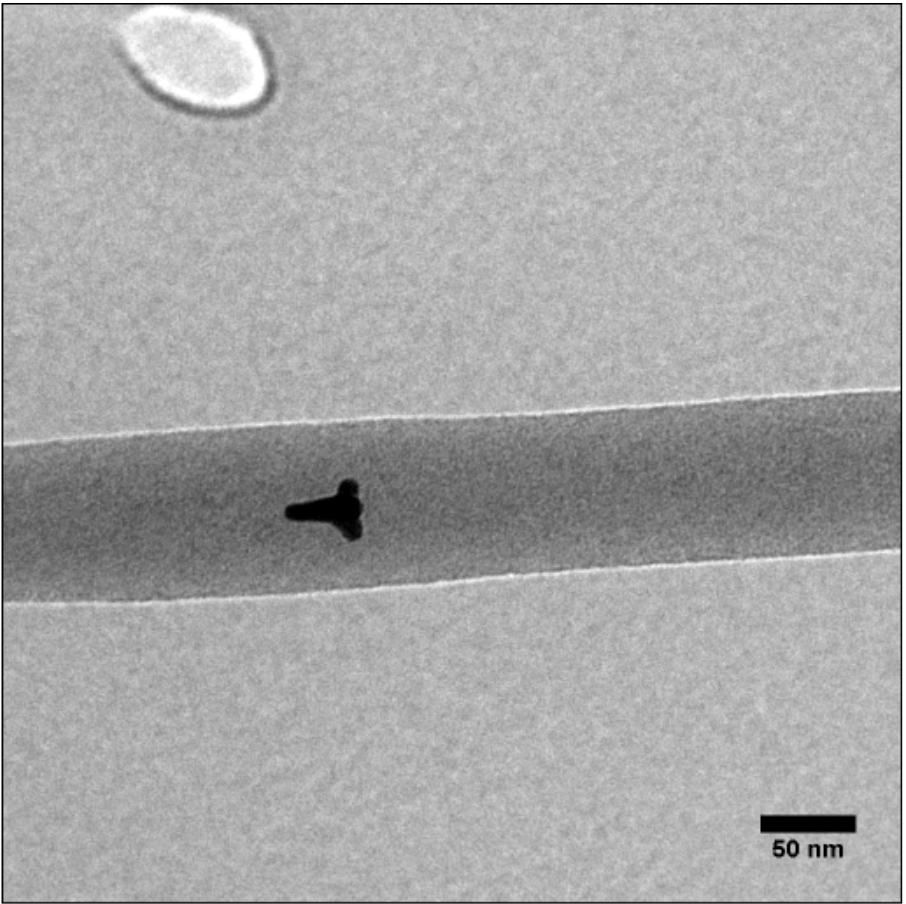
some nanomaterials I made: vertically aligned carbon nanofibers (left) and gold nanostars embedded in polymer nanofibers (right)